
KS-1 Novel Fertilizer-Drawn Forward Osmosis to Recover Nutrients from Source-Separated Wastewater and Urine for Urban Farming
Federico Volpin, Laura Chekli, Sherub Phuntsho and Ho Kyong Shon1*
School of Civil and Environmental Engineering, University of Technology Sydney (UTS),
City Campus, Broadway, NSW 2007, Australia
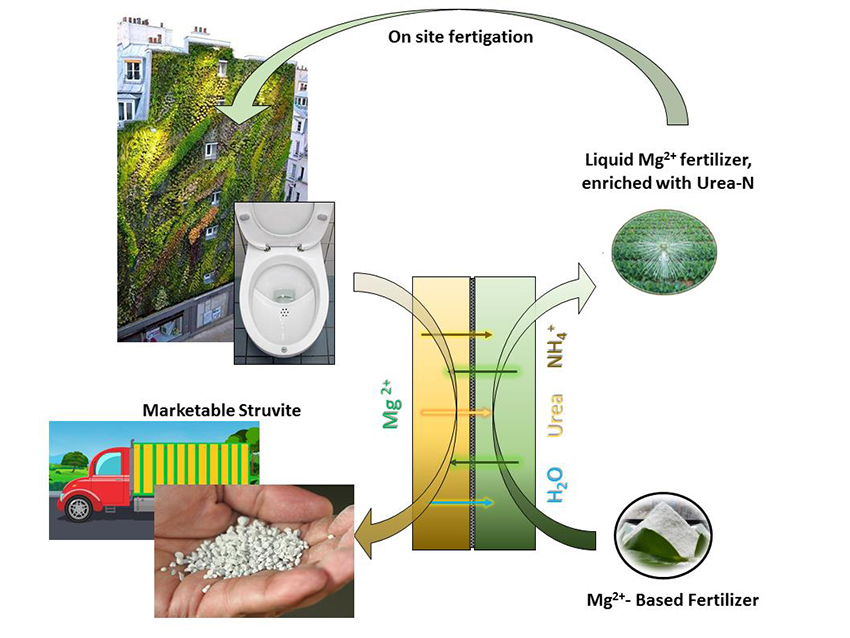 Figure 1 Schematic conceptualisation of the proposed presentation
Figure 1 Schematic conceptualisation of the proposed presentation
KS-2Controlling chloroplast performance: The role of the NTRC/2-Cys peroxiredoxin redox system
Francisco Javier Cejudo*
Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla and CSIC,
Avda Américo Vespucio 49, 41092-Sevilla, Spain

KS-3Identification of bioactive compounds responsible for therapeutic benefits of traditional Chinese medicine
Yang Ye*, Chunping Tang, Shuaizhen Zhou, Changqiang Ke
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica,
Chinese Academy of Sciences, Shanghai, 201203, China
Radix Stemonae (Baibu, 百部) have long been used as an antitussive and insecticidal agent in history. Three species, Stemona tuberosa, S. sessilifolia and S. japonica, are all documented in Chinese pharmacopoeia as plant origin of Baibu. More than 90 Stemona alkaloids featuring a pyrrolo[1,2-α]azepine or pyrido[1,2-α]azepine nucleus were characterized. The alkaloidal extracts and major alkaloids were found to exhibit significant antitussive activity in a citric acid-induced cough model. Stemofoline and its analogs demonstrated in vitro and in vivo insecticidal activity. A comprehensive investigation of three species collected from different habitats revealed that the alkaloidal constituents vary significantly with species and habitats while little with harvesting seasons. A multivariate statistical analysis based on the UPLC-QTof-MS was established to further evaluate chemical diversity of the Stemona plants. Chemical marker was revealed for each species through an OPLS-DA model. A PCA model based on the mass fingerprints was then built for identification and quality control of Baibu (Fig. 1).
Out study revealed that both crude alkaloidal extracts and different types of alkaloids exhibited strong antitussive activity while they also showed different side effects. S. sessilifolia growing in Chuzhou, recorded in ancient medicinal books as the geo-authentic species, contains a major alkaloid with the most potent antitussive activity and the least side effects. The findings provide scientific evidence for therapeutic applications of the TCM “Baibu” in history and in current clinic, and contribute to the global recognition of traditional Chinese medicines.
Keywords: Natural products; Radix Stemonae; Antitussive; Alkaloid
 Fig. 1. Score plot of a PCA model to distinguish S. tuberosa (red), S. sessilifolia (blue), and S. japonica (green).
Fig. 1. Score plot of a PCA model to distinguish S. tuberosa (red), S. sessilifolia (blue), and S. japonica (green).Acknowledgements
The authors thank the financial support from the Outstanding Young Scholar Grant, National Natural Science Foundation of China.

KS-4Microbial Ecology of Nitrogen Dynamics in Agricultural Soils: Key Drivers of Reductive N Transformation in Paddy Field and N2O Generation in Upland Field
Keishi Senoo1,2*
1 Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences,
2 Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Japan
Waterlogged paddy soils are characterized by development of anoxic zone, where reductive nitrogen transformations (RNT), i.e., denitrification, dissimilatory nitrate reduction to ammonium, and nitrogen fixation actively progress by soil microbes, leading to low leaching of nitrogen pollutants (nitrate and nitrous oxide gas) and large retention of nitrogen nutrition. Although many past studies have estimated diversity of RNT microbes based on PCR-based analyses of RNT genes, recent genomic researches have warned an underestimate of the diversity by previous methods.
To avoid such a risk, we performed shotgun sequencing analysis (metatranscriptomics) of soil RNA extracted from paddy soils using MiSeq sequencer. The sequences of RNT genes were retrieved from the metatranscriptomic libraries obtained and taxonomically annotated through a tandem similarity search with the BLAT and BLAST programs.
As a result, most of the RNT gene transcripts in paddy soils were derived from Deltaproteobacteria, particularly the genera Anaeromyxobacter and Geobacter. Despite the frequent detection of the rRNA of these microbes in paddy soils, their RNT-associated genes have rarely been identified in previous PCR-based studies. Anaeromyxobacter and Geobacter, well-known iron reducers universally dominating in paddy soils, were suggested to associate with RNT, partial reactions in denitrification, ammonium formation via nitrite reduction, and nitrogen fixation. By combination of these reactions, Anaeromyxobacter and Geobacter could generate ammonium, which contributes to nitrogen fertility of paddy soils.
Our metatranscriptomics provides novel insights into the diversity of RNT microbes in paddy soils and the ecological function of Deltaproteobacteria dominating in paddy soils, which may lead to the establishment of low-nitrogen agriculture.
Upland agricultural field is a major source of nitrous oxide (N2O), one of greenhouse gases and ozone depleting substances. N2O emission from agricultural fields is mostly derived from microbial transformation of nitrogen contained in fertilizers applied to soil. Information on the N2O generating soil microorganisms is essential to mitigate the N2O emission from agricultural fields.
We observed large emissions of N2O after basal and supplemental application of organic fertilizer to upland field soil. Soil microcosm study revealed that bacterial (basal application) and fungal (supplemental application) denitrification are largely responsible for the N2O emissions, whereas the contribution of nitrification is small. By combination of culture-dependent and culture-independent analysis, we successfully identified bacterial and fungal species responsible for the N2O emissions. Characterization of the N2O-generating microorganisms will be helpful to establish agricultural practice for N2O mitigation.

KS-5 Temperature-mediated responses in plants
Sibum Sung
Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, USA

KS-6Genomics-Based Enzyme Technology for Targeting the Sweet Spot
Dong-Woo Lee*
Dept. of Biotechnology, Yonsei University, Seoul 03722, Republic of Korea
